Ligustica Introgression pdf
|
|
|---|
|
Ligustica Introgression pdf |
Honeybee Genetics...
|
|---|
| Originally titled... | |
|
Genetic Analysis of Commercial Honey Bees (Hymenoptera: Apidae) from the Southeastern
United States. A paper by:- Nathan M. Schiff and Walter Sheppard(1) Bee Research Laboratory, Beltsville Agricultural Research Center-East, USDA-ARS, Building 476, Beltsville, MD 20705 Journal Economic Entomology 88(5): 1216 - 1220 (1995) | |
Approximately 3.2 million honey bee colonies are maintained by beekeepers in the United States and many are headed by commercially bred queens. We used mitochondrial DNA (mtDNA) and allozyme variation to characterize 142 breeder queen colonies from 22 apiaries in the southeastern United States that produced approximately 483,900 commercial honey bee queens in 1993. Analysis of mtDNA haplotypes showed that 4% of the 142 commercial breeder queen colonies were maternal descendants of Apis mellifera mellifera, a subspecies that was imported into the United States by the 17th century but is no longer used commercially. The other 96% were probably descendants of A. m. ligustica or A. m. carnica, subspecies imported in the 19th century which are still sold as commercial strains. Malate dehydrogenase allele frequencies for the 142 breeder queen colonies were determined to be Mdh65 = 0.50, Mdh80 = 0.23, Mdh100 = 0.27. Five other enzymes known to be polymorphic in adult honey bees were invariant.
Significant genetic differences between commercial and feral populations suggest that the feral population may represent a novel source of genetic variation for breeding programs.
The honey bee, Apis mellifera L., is not native to the New World. It is endemic to Europe, Africa, and the Middle East where it has evolved into 25 recognizable geographic races or subspecies (Ruttner 1992). Eight subspecies are known to have been introduced to North America (Sheppard 1989). A 9th subspecies was introduced to Brazil in 1957 and its descendants, known as Africanized honey bees, spread to the United States in 1990.
The current honey bee population of the United States is composed of both commercial and feral sub populations. The commercial subpopulation is actively maintained in hives by beekeepers to produce honey and other bee products, and for pollination of various crops. Beekeepers typically replace the queen every 1 to 2 years, to ensure good egg production and colony vigour, often with replacement queens purchased from queen breeders. Honey bee queen breeders commonly offer commercial strains of A. m. carnica Pollman, A. m. ligustica Spinola, and A. m. caucasica Gorbatschev for sale, thus, the roughly 3.2 million colonies maintained by beekeepers in the United States (Anonymous 1993) may be descended primarily from these subspecies. Replacement queens that head commercial colonies are the daughters of relatively few selected breeder queens that are used excusively for queen production. The production of replacement queens from a limited number of breeder queens creates the potential for reduction of genetic variability. The size of the breeding population is unknown, however, and several studies of commercial colonies have found a range of Mdh allele frequencies (Sylvester 1976, Nunamaker 1980, Page and Metcalf 1988, Sheppard 1988, Spivak et al. 1988, Hung et al. 1991).
The feral subpopulation consists of colonies not actively maintained by beekeepers with queen replacement. These colonies occupy a variety of natural and artificial nesting sites and likely represent a wider mixture of the introduced races. The extent of gene flow between feral and commercial honey bee populations is unknown, but considering the unknown subspecies contribution to the 2 groups, the selective pressures exerted by bee breeders on the commercial population, and unknown selective pressures on the feral population, there is potential for genetic differentiation. Sheppard (1988) found significant Mdh allele differences between 14 feral (as defined in the current study) and 25 commercial colonies.
Recently 692 feral colonies from the southern United States were characterized on the basis of mtDNA haplotypes and allozyme variation (Schiff and Sheppard 1993, Schiff et al. 1994). In the current study, we used mtDNA and enzyme variation to genetically characterize breeder queen colonies from the southeastern United States. Results are compared with previously collected data from feral colonies from this area to assess differentiation between the 2 groups.
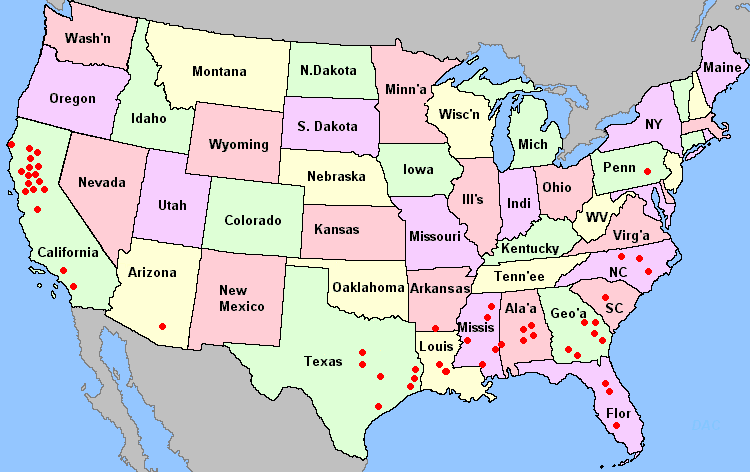
Fig. 1. Distribution of commercial queen breeders in the United States.
A list of commercial queen producers in the United States compiled from recent advertising literature revealed 2 major queen production areas, the southeastern United States and California (Fig. 1). To sample a large proportion of the commercial gene pool we chose to sample the breeder queen colonies. Samples of workers from 185 colonies belonging to 22 queen producing apiaries from the southeastern United States were collected. Of these, 142 were breeder queen colonies and 43 were colonies headed by daughters of breeder queens. The 43 daughter colonies were analyzed for comparison with breeder queen colonies and to increase the probability of detecting rare alleles or haplotypes. In a few cases, the breeder queen colonies at an apiary were unmarked. In these instances, we used a single colony known to be headed by a daughter of a breeder queen to represent the apiary, even though other colonies sampled probably represented additional breeder queens from that apiary. Adult workers were collected directly from hives, and the bees were frozen and kept in liquid nitrogen until storage at -80°C in the laboratory. Data on the number of breeder queens used in a year and number of marketed queens were collected from each bee breeder.
Mitochondrial DNA analysis consisted of total nucleic acid extraction (Sheppard and McPheron 1991) from 2 worker thoraces followed by digestion with EcoRI according to manufacturer instructions. The products were separated on 1% agarose gels, visualized with ethidium bromide and photographed for documentation, If all lanes were not easily scorable, nucleic acids were transferred to a nylon membrane using a capillary blot and probed using previously reported methods (Schiff et al. 1994).
Horizontal starch gel electrophoresis was performed on a single worker bee thorax for each of the 185 colonies using previously described methods (Sheppard and Berlocher 1984, 1985; Del Lama et al. 1988). A single worker per colony was used to prevent over representation of the genotype from a particular queen or drone. Six enzyme systems known to be polymorphic in the adult honey bee were examined. These were malate dehydrogenase (MDH-1), phosphoglucomutase (PGM), hexokinase (HK), aconitase (ACON-1), malic enzyme (ME), and esterase (EST-3) (Mestriner and Contel 1972; Sheppard and Berlocher 1984, 1985; Sheppard and McPheron 1986; Del Lama et al. 1988). Mendelian inheritance of electromorphs was assumed, although this has not been established for ME, PGM, or ACON-1.
Counts of mtDNA haplotypes and Mdh alleles were compared between feral and commercial populations using chi-square analyses. Direct comparison of Mdh alleles from this study was possible with those of the feral populations of Schiff et al. (1994) because a single worker from each colony was determined in both cases. To compare bee allele frequencies with those of previous studies, which based allele frequencies on multiple workers per colony, we multiplied reported allele frequencies by 2 times the number of colonies studied to estimate counts of alleles for chi-square analysis.
The 22 queen producing apiaries studied used a total of 308 breeder queens to produce approximately 483,900 marketable queens (an average of 1,571 queens per breeder queen). This represents replacement queens for about 1/6 of the estimated 3.2 million colonies actively maintained in the United States.
Several different kinds of honey bees were advertised for sale by the queen breeders, including commercial strains of 3 of the subspecies originally introduced to the United States: Italians, Caucasians, and Carniolans. Additionally, 3 strains selected for beekeeping were sold: 'Buckfast', selected from crosses of various subspecies by a bee breeder in England; 'Midnight', derived from Caucasian bees; and 'Starline', derived from Italian stock. Marketable queens of these strains produced by the 22 apiaries were 393,400 Italians, 30,000 Caucasians, 3,500 Carniolans, 25,000 Buckfast, 12,000 Midnight, and 33,000 Starline. Although Caucasian and Buckfast samples were large enough for statistical analysis, they were not significantly different than the population as a whole and all strains were treated together as commercial bees.
Two mtDNA haplotypes were detected in the 142 breeder queen colonies analyzed. Six colonies from 3 of the apiaries had the haplotype associated with A. m. mellifera or A. m. iberica Goetze (Smith et al. 1991). These 3 apiaries produced a total of 50,000 marketable queens and, assuming that all breeder queens within an apiary produced an equal proportion of the total queens sold, the 6 A. m. mellifera haplotypes produced 14,000 (3%) of the queens sold by the 22 apiaries. The remaining 136 breeder queens all had mtDNA haplotypes associated with A. m. carnica and A. m. ligustica (Table 1.) and accounted for 433,900 (97%) of the total queens sold. This is significantly different (X2=63.1, P<0.001) than the feral population of the southern United States, where 36.7% of 692 feral colonies had the A. m. mellifera/iberica haplotype (Schiff et al. 1994). The lack of A. m. mellifera haplotypes in the commercial population is indicative of restricted gene flow between feral and commercial populations. Until A. m. ligustica was introduced by bee breeders in 1859, A. m. mellifera was the only subspecies present in the United States. Gene flow between commercial and feral populations likely has occurred through swarming and open matings since that time. However, the maternally inherited mtDNA of A. m. mellifera has made little intrusion into commercial populations, demonstrated by the low frequency of iA. m. mellifera/i mtDNA haplotypes in this group (3%). Perhaps, through selection or other breeding practices, bee breeders have contributed to this asymmetry in mtDNA haplotype frequencies. A 3rd haplotype, A. m. lamarckii, which was present in 2% of feral colonies (Schiff et al. 1994) was not found in the sampled breeder queen population.
Table 1. MDH allele frequencies mtDNA haplotypes for the U.S. commercial breeder queen population.
| Designation | No. of colonies | Allele designations | Haplotypes | |||
|---|---|---|---|---|---|---|
| Mdh65 | Mdh80 | Mdh100 | mellifera /iberica | carnica/ ligustica | ||
| Breeders | 142 | 0.50 | 0.23 | 0.27 | 6 | 136 |
| Daughters | 43 | 0.51 | 0.16 | 0.33 | 2 | 41 |
Enzyme polymorphism in commercial honey bees was lower than in the feral population. Of the 6 enzymes known to be polymorphic in adult honey bees, only MDH was polymorphic in the commercial colonies of this study, whereas uncommon alleles for ME, EST 3, and PGM were detected in feral colonies (Schiff et al. 1994). Although it may be argued that the differences in sample sizes may be responsible for the higher monomorphism of the commercial group, it is relevant to note that both commercial and feral populations had fewer alleles than were reported from 23 A. mellifera colonies from Europe (Sheppard 1988).
Malate dehydrogenase allele frequencies are reported for the breeder queen population in Table 1. They were significantly different than those found in feral colonies (Schiff et al. 1994) (X2 = 13.1, df = 2, P<0.01). The feral U.S. population has a higher Mdh80 allele frequency, which might be expected because this allele has been associated with A. m. mellifera (Badino et al. 1984, Sheppard and Berlocher 1984, Cornuet et al. 1986) and the A. m. mellifera mtDNA haplotype is common in the feral population (Schiff et al. 1994). Populations of A. m. ligustica from Italy and A. m. carnica from Austria and Yugoslavia had a combined Mdh80 allele frequency of 0.03 (694 colonies combined from Badino et al. 1983, 1984; Sheppard and Berlocher 1985; Comparini and Biasiolo 1991), significantly lower than our samples of U.S. breeder queens (Mdh80 allele frequency of 0.23; X2 = 76.4, df = 1, P<0.001). This may be indicative of paternal gene flow from the feral to the breeder queen population, which would be undetected in the analysis of mtDNA. The Mdh100 allele frequency of the breeder queens was 0.27. This allele has been reported to be common in African races of honey bees (Sylvester 1982, Nunamaker et al. 1984, Cornuet et al. 1986) and some of the European subspecies that form the basis of the U.S. commercial population (Badino et al. 1983, Sheppard and Berlocher 1984, Sheppard and McPheron 1986, Badino et al. 1988, Comparini and Biasiolo 1991). MDH allele frequencies of the breeder queen populations were also significantly different than those previously reported for feral populations by Sheppard (1988) (X2 = 5.8, df = 2, P<0.05).
Table 2. MDH allele frequencies for feral and commercial populations of honey bees
| Designation | No. of colonies | Allele designations | Reference | ||
|---|---|---|---|---|---|
| Mdh65 | Mdh80 | Mdh100 | |||
| Feral A(a) | 692 | 0.47 | 0.33 | 0.20 | 1 |
| Feral B(a) | 14 | 0.40 | 0.42 | 0.18 | 2 |
| Commercial A | 5 | 0.55 | 0.03 | 0.42 | 3 |
| Commercial B(a) | 74 | 0.39 | 0.37 | 0.24 | 4 |
| Commercial C(a) | 24 | 0.72 | 0.06 | 0.22 | 5 |
| Commercial D | 25 | 0.62 | 0.08 | 0.30 | 2 |
| Commercial E | 10 | 0.46 | 0.20 | 0.34 | 6 |
| Commercial F(a) | 20 | 0.77 | 0.08 | 0.16 | 7 |
| Total A-F | 158 | 0.53 | 0.22 | 0.25 | 2-7 |
References (table 2): 1, Schiff et al. 1994; 2, Sheppard 1988; 3, Spivak et al. 1988; 4, Nunamaker 1980; 5, Sylvester 1976; 6, Hung et al. 1991; 7, Page and Metcalf 1988.
Items marked (a) Significantly different than commercial population of this study (X2 test, P < 0.05).
Malate dehydrogenase allele frequencies for commercial colonies from 6 previous studies were extremely variable (Table 2), but when they were combined the overall mean MDH allele frequencies were not significantly different than those reported for the commercial breeder queen population in this study (X2 = 0.45, df = 2, P > 0.5). Individually, allele frequencies reported for commercial bees in some studies were significantly different than those found in this study (Nunamaker 1980, X2 = 9.8, df = 2, P < 0.01; Sylvester 1976, X2 = 8.9, df = 2, P < 0.05; Page and Metcalf 1988, X2 = 10.6, df = 2, P < 0.01). This may result from differences in sample size, sampling methods, or a combination of both when comparing the current with previous studies. Weighting the breeder queen contributions by the proportion of marketable queens each apiary produced, provided another estimate of MDH frequencies for the commercial population. However, the frequencies (Mdh65 = 0.48, Mdh80 = 0.20, Mdh100 = 0.32) were not significantly different than those measured for the breeder queen population (Table 1, X2 = 2.27, df = 2, P < 0.10).
To test for heterogeneity within the breeder queen population, we compared MDH allele frequencies for 2 subsamples with those of the total population. Georgia, with 35 colonies, was the eastern subsample and Texas, with 39 colonies, the western subsample. Neither subsample differed significantly from the total population (chi-squares of 1.6 and 3.1, df = 2, P > 0.1, respectively). In contrast, when the feral population was divided by states, several subpopulations were significantly different than the feral population as a whole (Schiff et al. 1994). This suggests that the commercial population is more homogeneous than the feral population, which might be expected because of the potential for queen production methods to reduce the commercial gene pool. Analysis of the 43 additional commercial colonies that were collected from breeder queen apiaries supported the idea of homogeneity among the commercial gene pool. Mitochondrial DNA analysis revealed 2 additional A. m. mellifera haplotypes and 41 A. m. carnica/A. m. ligustica haplotypes. Mdh allele frequencies were not significantly different from the breeder queen population (X2 = 1.9, df = 2, P > 0.1), and no additional polymorphism was detected for any of the enzymes screened.
Based on mtDNA haplotypes and allozyme variability we report significant differences between feral and commercial populations of honey bees from the southern United States. Subsamples of the commercial breeder queen population were not significantly different from the total population, indicating less heterogeneity within this population than previously reported for feral honey bees. The significance of these results lies in the context in which honey bee queens are produced and distributed within the United States. Virgin queens are produced from breeder queens and then mated by local drones derived primarily from colonies located in a mating apiary. The relatively few (308) breeder queens used to produce annual replacement queens for about 1/6 of the U.S. commercial population certainly represents a genetic bottleneck. Further study may reveal whether the genetic differentiation between commercial and feral populations can be exploited to improve bee breeding. Our results suggest that the potential to select for desirable traits may be enhanced by including feral colonies in the screening effort.
We thank the following people for helping us collect samples of honey bees: L. Busby, G. Curtis, D. Drew, W. Gafford, M. Hardeman, R. Harrell, L. Hines, H. Homan, C. Lester, C. Linkous, G. Loper, G. McCary, O. Mitchell, T. Norman, F. Rossman, J. Shumans, S. Spell, G. Waller, B. Weaver, M. Weaver, A. Webb, R. Wilbanks, K. Williams, and H. York. We also thank B. A. McPheron and S. Berlocher for reviewing the manuscript. This work was supported in part by NRICGP Grant number 9301929.
Anonymous. 1993. Honey. USDA Agricultural Statistics Board, National Agricultural Statistics Service. Washington DC.
Badino, G., G. Celebrano, and A. Manino. 1983. Population structure and Mdh-1 locus variation in Apis mellifera ligustica. J. Hered. 74: 443-446.
1984. Population genetics of Italian honeybee (Apis mellifera ligustica Spin.) and its relationships with neighbouring subspecies. Boll. Mus. Reg. Sci. Nat. Torino 2: 571-584.
Badino, G., G. Celebrano, A. Manino, and M. D. Ifantidis. 1988. Allozyme variability in Greek honeybees (Apis mellifera L.). Apidologie 19: 377-386.
Comparini, A. and A. Biasiolo. 1991. Genetic discrimination of Italian bee, Apis mellifera ligustica versus Carniolan bee, Apis mellifera carnica, by allozyme variability analysis. Biochem. Syst. Ecol. 19: 189-194.
Cornuet, J. M., A. Daoudi, and C. Chevalet. 1986. Genetic pollution and number of matings in a black honey bee (Apis mellifera mellifera) population. Theor. Appl. Genet. 73: 223-227.
Del Lama, M. A., R. A. Figueiredo, A.E.E. Soares, and S. N. Del Lama. 1988. Hexokinase polymorphism in Apis mellifera and its use for Africanized honeybee identification. Rev. Brazil. Genet. 11: 287-297.
Hung, A. C. F., W. L. Rubink, and R. M. Crewe. 1991. Association of enzyme/protein loci in individual honey bees (Apis mellifera L.) (Hymenoptera: Apidae). Bee. Sci. 1: 225-229.
Mestriner, M. A., and E.P.B. Contel. 1972. The P-3 and EST loci in the honeybee Apis mellifera. Genetics 72: 733-738
Nunamaker, R. A. 1980. Subspecies determination in the honey bee (Apis mellifera L.) based on isoelectric focusing of malate dehydrogenase. PhD dissertation. University of Wyoming, Laramie.
Nunamaker, R. A., W. T. Wilson, and B. E. Haley. 1984. Electrophoretic detection of Africanized honey bee (Apis mellifera scutellata) in Guatemala and Mexico based on malate dehydrogenase allozyme patterns. J. Kans. Entomol. Soc. 57: 622-631.
Page, R. E., and R. A. Metcalf. 1988. A population estimate of MDH allozyme frequencies for the honey bee, Apis mellifera L. (Hymenoptera: Apidae). Pan. Pac. Entomol. 64: 285-289.
Ruttner, F. 1992. Naturgeschichte der Honigbienen. Ehrenwirt, Munich.
Schiff, N. M., and W. S. Sheppard. 1993. Mitochondrial DNA evidence for the 19th century introduction of African honey bees into the United States. Experientia 49: 350-352.
Schiff, N. M., and W. S. Sheppard, G. M. Loper, and H. Shimanuki. 1994. Genetic diversity of feral honey bee (Hymenoptera: Apidae) populations in the southern United States. Ann. Entomol. Soc. Am. 87: 842-848.
Sheppard, W. S. 1988. Comparative study of enzyme polymorphism in United States and European honey bee (Hymenoptera: Apidae) populations. Ann. Entomol. Soc. Am. 81: 886-889.
1989. A history of the introduction of honey bee races into the United States. Am. Bee J. 129: 617-619, 664-667.
Sheppard, W. S., and S. H. Berlocher. 1984. Enzyme polymorphisms in Apis mellifera mellifera from Norway. J. Apic. Res. 23: 64-69.
1985. New Allozyme variability in Italian honey bees, Apis mellifera ligustica. J. Hered. 76: 45-48.
Sheppard, W. S., and B. A. McPheron. 1986. Genetic variation in honey bees from an area of racial hybridization in western Czechoslovakia. Apidologie 17: 21-32.
1991. Ribosomal DNA diversity in Apidae, pp 89-102 In D. R. Smith [ed.], Diversity in the genus Apis. Westview, Oxford.
Smith, D. R., M. F. Palopoli, B. R. Taylor, L. Garnery, J. M. Cornuet, M. Solignac, and W. M. Brown. 1991. Geographical overlap of two mitochondrial genomes in Spanish honeybees (Apis mellifera iberica). J. Hered. 82: 96-100.
Spivak, M., T. Ranker, O. Taylor, Jr., W. Taylor, and L. Davis. 1988. Discrimination of Africanized honey bees using behavior, cell size, morphometrics, and a newly discovered isozyme polymorphism, pp 313-324. In G. R. Needham, R. E. Page, Jr., M. Delfinado-Baker, and C. E. Bowman [eds.], Africanized honey bees and bee mites. Ellis Horwood, Chichester.
Sylvester, H. A. 1976. Allozyme variation in honey bees (Apis mellifera L.). PhD dissertation. University of California, Davis.
1982. Electrophoretic identification of Africanized honeybees. J. Apic. Res. 21: 93-97.
Received for publication 31 August 1994; accepted 28 April 1995.
Dave Cushman's Note...
The diagram has been redrawn and may not be quite as precise in the positioning of queen
rearers as the original, but any error is small and does not affect the basic geographic
distribution. Typos and some American spellings have been corrected, the subtitling...
"Systematic elimination of the wrong portions of hybridised genes" is my personal
spin and comment as a breeder of A. m. mellifera types of bee that have largely been eliminated from the US gene pool
(in my opinion to the detriment of future selection). Although biased in favour of A. m. mellifera bee
types myself, the authors are reporting on the status quo and not advocating such
elimination, I can recommend the reference list for further reading. D.A.C.
Dave Cushman.
Page created March 2004
Page updated 18/12/2022
Printed from Dave Cushman's website Live CD version
Originated... March;2004, Revised... 22 Sepember 2004, Upgraded... 16 Sepember 2007, Robotic Changes... 07 December 2007,
|